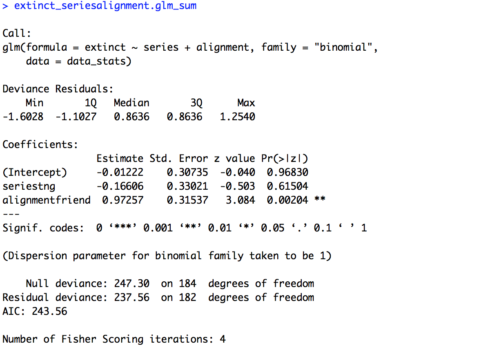
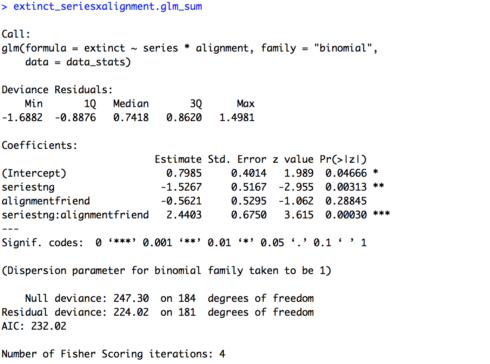
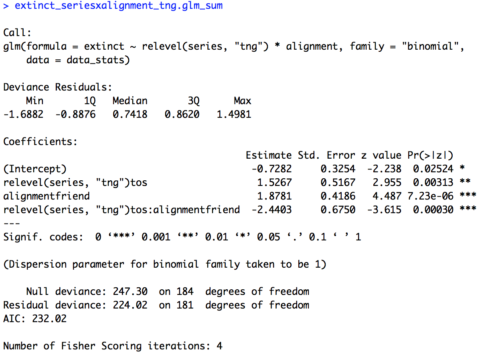
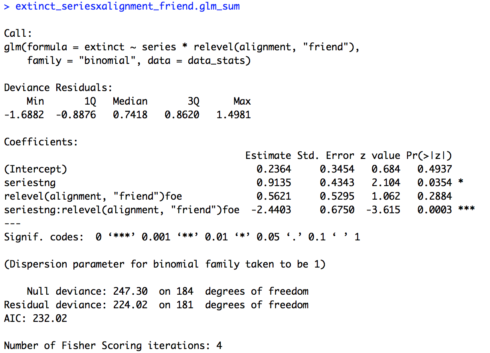
In today’s lesson we’ll learn about linear mixed effects models (LMEM), which give us the power to account for multiple types of effects in a single model. This is Part 1 of a two part lesson. I’ll be taking for granted some of the set-up steps from Lesson 1, so if you haven’t done that yet be sure to go back and do it.
By the end of this lesson you will:
- Have learned the math of an LMEM.
- Be able to make figures to present data for LMEMs.
- Be able to run some (preliminary) LMEMs and interpret the results.
Note, we won’t be making an R Markdown document today, that will be saved for Part 2. There is a video in end of this post which provides the background on the math of LMEM and introduces the data set we’ll be using today. There is also some extra explanation of some of the new code we’ll be writing. For all of the coding please see the text below. A PDF of the slides can be downloaded here. Before beginning please download these text files, it is the data we will use for the lesson. Note, there are both two text files and then a folder with more text files. Data for today’s lesson comes from actual students who took the in person version of this course. They participated in an online version of the Stroop task. All of the data and completed code for the lesson can be found here.
Lab Problem
Unlike past lessons where our data came from some outside source, today we’ll be looking at data collected from people who actually took this course. Participants did an online version of the Stroop task. In the experiment, participants are presented color words (e.g. “red”, “yellow”) where the color of the ink either matches the word (e.g. “red” written in red ink) or doesn’t match the word (e.g. “red” written in yellow ink). Participants have to press a key to say what the color of the ink is, NOT what the text of the word is. Participants are generally slower and less accurate when the there is a mismatch (incongruent trial) than when there is a match (congruent trial). Furthermore, we’re going to see how this effect may change throughout the course of the experiment. Our research questions are below.
- Congruency: Are responses to incongruent trials less accurate and slower than to congruent trials?
- Experiment half: Are responses more accurate and faster in the second half of the experiment than the first half of the experiment?
- Congruency x Experiment half: Is there an interaction between these variables?
Setting up Your Work Space
As we did for Lesson 1 complete the following steps to create your work space. If you want more details on how to do this refer back to Lesson 1:
- Make your directory (e.g. “rcourse_lesson6”) with folders inside (e.g. “data”, “figures”, “scripts”, “write_up”).
- Put the data files for this lesson in your “data” folder, keep the folder “results” intact. So your “data” folder should have the results folder and the two other text files.
- Make an R Project based in your main directory folder (e.g. “rcourse_lesson6”).
- Commit to Git.
- Create the repository on Bitbucket and push your initial commit to Bitbucket.
Okay you’re all ready to get started!
Cleaning Script
Make a new script from the menu. We start the same way we usually do, by having a header line talking about loading any necessary packages and then listing the packages we’ll be using. We’ll be using both dplyr and purrr. Copy the code below to your script and and run it.
## LOAD PACKAGES ####
library(dplyr)
library(purrr)
We’ll start by reading our data in the “results” folder. In this folder is a separate text file for each of the subjects* who completed the experiment, with all of their responses to each trial. We’ll read in all of these files at once using purrr. For a reminder on how this code works see Lesson 4. Copy and run the code below to read in the files.
## READ IN DATA
# Read in full results
data_results = list.files(path = "data/results", full.names = T) %>%
map(read.table, header = T, sep="\t") %>%
reduce(rbind)
Take a look at “data_results”. You’re probably thinking that it’s very messy, and with far more columns than are probably necessary for our analysis. One of my goals for this lesson is to show you what real, messy data looks like. Sometimes you can’t control the output of your data from certain experimental programs, and as a result you can get data frames like this. We’ll work on cleaning this up in a minute.
In addition to reading in our results data we also have our two other files to read in. In the “rcourse_lesson6_data_subjects.txt” file is some more information about each of our subjects, such as their sex and if their native language is French or not (the experiment was conducted in French). The “rcourse_lesson6_data_items.txt” file has specific information about each item in the experiment, such as the word and the ink color. Copy and run the code below to read in these single files.
# Read in extra data about specific subjects
data_subjects = read.table("data/rcourse_lesson6_data_subjects.txt", header=T, sep="\t")
# Read in extra data about specific items
data_items = read.table("data/rcourse_lesson6_data_items.txt", header=T, sep="\t")
Now we can start cleaning our data including getting rid of some of our unnecessary columns. I’m going to start with our results data. We’re going to start be renaming a bunch of the columns so that they are more readable using the rename() call in dplyr. To learn more about how this call works watch the video. Copy and run the code below.
## CLEAN DATA ####
# Fix and update columns for results data, combine with other data
data_clean = data_results %>%
rename(trial_number = SimpleRTBLock.TrialNr.) %>%
rename(congruency = Congruency) %>%
rename(correct_response = StroopItem.CRESP.) %>%
rename(given_response = StroopItem.RESP.) %>%
rename(accuracy = StroopItem.ACC.) %>%
rename(rt = StroopItem.RT.)
Now my results file has much easier to understand column names, and it matches my common pattern of only using lower case letters and a “_” to separate words. However, I still have a bunch of other columns in the data frame that I don’t want. To get rid of them I’m going to use the select() call to only include the specific columns I need. Watch the video to learn more about this code. Copy and run the updated code below.
data_clean = data_results %>%
rename(trial_number = SimpleRTBLock.TrialNr.) %>%
rename(congruency = Congruency) %>%
rename(correct_response = StroopItem.CRESP.) %>%
rename(given_response = StroopItem.RESP.) %>%
rename(accuracy = StroopItem.ACC.) %>%
rename(rt = StroopItem.RT.) %>%
select(subject_id, block, item, trial_number, congruency,
correct_response, given_response, accuracy, rt)
Now that my messy results data is cleaned up, I’m going to combine it with my other two data frames with an inner_join() call, so that now I’ll have all of my information, results, subjects, items, in a single data frame. Copy and run the updated code below.
data_clean = data_results %>%
rename(trial_number = SimpleRTBLock.TrialNr.) %>%
rename(congruency = Congruency) %>%
rename(correct_response = StroopItem.CRESP.) %>%
rename(given_response = StroopItem.RESP.) %>%
rename(accuracy = StroopItem.ACC.) %>%
rename(rt = StroopItem.RT.) %>%
select(subject_id, block, item, trial_number, congruency,
correct_response, given_response, accuracy, rt) %>%
inner_join(data_subjects) %>%
inner_join(data_items)
There’s still one more thing I’m going to do before I finish with this data frame. Right now there is a column called “block”. If you run an xtabs() call on “block” you’ll see that there are four of them in total. This is because in the experiment I had eight unique items repeated four times in a randomized order within four blocks. However, our research question asks about experiment half, not experiment quarter. So I’m going to make a new column called “half” that codes if a given trial occurred in the first or second half of the experiment. I’ll do this with a mutate() call and an ifelse() statement. Copy and run the final updated code below.
data_clean = data_results %>%
rename(trial_number = SimpleRTBLock.TrialNr.) %>%
rename(congruency = Congruency) %>%
rename(correct_response = StroopItem.CRESP.) %>%
rename(given_response = StroopItem.RESP.) %>%
rename(accuracy = StroopItem.ACC.) %>%
rename(rt = StroopItem.RT.) %>%
select(subject_id, block, item, trial_number, congruency,
correct_response, given_response, accuracy, rt) %>%
inner_join(data_subjects) %>%
inner_join(data_items) %>%
mutate(half = ifelse(block == "one" | block == "two", "first", "second"))
Normally we’d stop here with our cleaning, but since we’re working with reaction time data there’s a few more things we need to do. It’s common when working with reaction time data to drop data points that have particularly high or low durations. How people do this can vary across different researchers and experiments. We’ll be using one particular method today, but I want to stress that I’m by no means saying this is the best or only way to do outlier removal, it’s simply the method we’ll be using today as an example.
Before I remove my outliers I’m going to summarise my data to see what qualifies something to be an outlier. To do this I’m going to make a new data frame called “data_rt_sum” based on our newly made “data_clean”. I’m going to compute unique outlier criteria for each subject and within each of my experimental variables, congruency and experiment half. So I’m going to use a group_by() call before computing my outliers. Again, some people choose to compute outliers across the whole data set not grouping by anything, or only grouping by some other variables. Now that my data is grouped I’m going to summarise() it to get the mean reaction time (“rt_mean”) and the standard deviation (“rt_sd”). Finally, I’ll ungroup() the data when I’m doing summarising. Take a look at the code below and when you’re ready go ahead and run it.
# Get RT outlier information
data_rt_sum = data_clean %>%
group_by(subject_id, congruency, half) %>%
summarise(rt_mean = mean(rt),
rt_sd = sd(rt)) %>%
ungroup()
Take a look at the data frame. You should have 112 rows, 28 subjects each with 4 data points (1) congruent – first half, 2) incongruent – first half, 3) congruent – second half, 4) incongruent – second half). You should also have our newly created columns that give you the mean reaction time and the standard deviation for each grouping. We’re not quite done though. For my outlier criterion I want to say that any reaction time two standard deviations above or below the mean is considered an outlier. To do this I’m going to start by making two new columns that specify exactly what those values are, “rt_high” for reaction times two standard deviations above the mean, and “rt_low” for reaction times two standard deviations below the mean. Copy and run the updated code below when you’re ready.
data_rt_sum = data_clean %>%
group_by(subject_id, congruency, half) %>%
summarise(rt_mean = mean(rt),
rt_sd = sd(rt)) %>%
ungroup() %>%
mutate(rt_high = rt_mean + (2 * rt_sd)) %>%
mutate(rt_low = rt_mean - (2 * rt_sd))
Now I want to remove any data points where the reaction time is above or below my threshold. At this time I’m also going to make two separate data frames, one for the accuracy analysis and one for the reaction time analysis. I’ll start with the data frame for my accuracy analysis. Note, sometimes people don’t remove data points from accuracy analyses based on reaction time data, but include all data points no matter what. Again, I’m not saying that the method we’re using here is the best or only way to run the analysis, it’s just the one we’ll be using for demonstration. To do this I’m first going to join my new data frame “data_rt_sum” with my previously cleaned up data frame, “data_clean”. This will add my new columns (“rt_mean”, “rt_sd”, “rt_high”, “rt_low”) as appropriate for each data point. Next, I’m going to use two filter() calls to drop any data points where reaction times are above my “rt_high” or below my “rt_low”. Copy and run the code below.
# Remove data points with slow RTs for accuracy data
data_accuracy_clean = data_clean %>%
inner_join(data_rt_sum) %>%
filter(rt = rt < rt_high) %>%
filter(rt = rt > rt_low)
I’m also going to make my data frame for the reaction time analysis. I’m going to use my newly made “data_accuracy_clean” data frame as a base, since that one already has my reaction time outliers dropped. However, I’m also going to now drop any data points where the subject gave an incorrect response, as for my reaction time analysis I only want to look at correct responses; I’ll do this with a filter() call. Copy and run the code below.
# Remove data points with incorrect response for RT data
data_rt_clean = data_accuracy_clean %>%
filter(accuracy == "1")
Before we move to our figures script be sure to save your script in the “scripts” folder and use a name ending in “_cleaning”, for example mine is called “rcourse_lesson6_cleaning”. Once the file is saved commit the change to Git. My commit message will be “Made cleaning script.”. Finally, push the commit up to Bitbucket.
Figures Script
Make a new script from the menu. You can close the cleaning script or leave it open, but we will be coming back to it. This new script is going to be our script for making all of our figures. We’ll start by using our source() call to read in our cleaning script, and then we’ll load our packages, in this case ggplot2 and RColorBrewer. For a reminder of what source() does go back to Lesson 2. Assuming you ran all of the code in the cleaning script there’s no need to run the source() line of code, but do load the packages. As I mentioned, we’ll also be using a new package called RColorBrewer. If you haven’t used this package before be sure to install it first using the code below. Note, this is a one time call, so you can type the code directly into the console instead of saving it in the script.
install.packages("RColorBrewer")
Once you have the package installed, copy the code below to your script and and run it as appropriate.
## READ IN DATA ####
source("scripts/rcourse_lesson6_cleaning.R")
## LOAD PACKAGES ####
library(ggplot2)
library(RColorBrewer)
Now we’ll clean our data specifically for our figures. Remember we have two different analyses so we’ll have two separate data frames for our figures, one for accuracy and one for reaction times. I’ll start with my accuracy data and I’m going to get it ready to make some boxplots. Since right now my dependent variable is all “0”s and “1”s I’m going to need to summarise to get a continuous dependent variable that I can plot. I’ll do this by summarising over subject and each of the independent variables. All of this should be familiar to you from past lessons with logistic regression. In addition to summarising my data for the figure, I’m also going to change my names of the levels for the “congruency” variable using a “mutate()” call. Copy and run the code below.
## ORGANIZE DATA ####
# Accuracy data
data_accuracy_figs = data_accuracy_clean %>%
group_by(subject_id, congruency, half) %>%
summarise(perc_correct = mean(accuracy) * 100) %>%
ungroup() %>%
mutate(congruency = factor(congruency, levels = c("con", "incon"),
labels = c("congruent", "incongruent")))
Let’s start now by making the boxplot for the accuracy data. At this point all of the code should be familiar to you. Copy and run the code to make the figure and save it to a PDF.
## MAKE FIGURES ####
# Accuracy figure
accuracy.plot = ggplot(data_accuracy_figs, aes(x = half, y = perc_correct,
fill = congruency)) +
geom_boxplot() +
ylim(0, 100) +
geom_hline(yintercept = 50)
pdf("figures/accuracy.pdf")
accuracy.plot
dev.off()
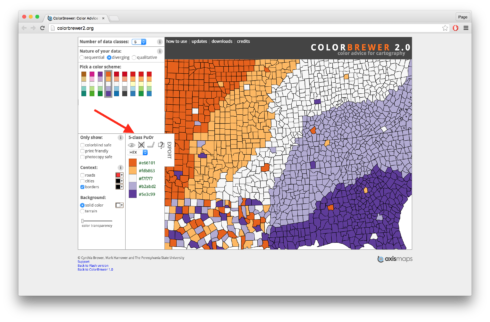
One thing we didn’t do was set our colors. In previous lessons we used scale_fill_manual() and then wrote the names for the colors we wanted to use. Today we’re going to use the package RColorBrewer. RColorBrewer has a series of palettes of different colors. To decide which palette to use I often use the website Color Brewer 2.0. With this site you can set the number of colors you want with the “Number of data classes” menu at the top. There are also different types of palettes depending on if you want your colors to be sequential, diverging, or qualitative. For our figures I’m going to set my “Number of data classes” to 5 and go with a diverging palette. Specifically, I’m going to use the one where the top is orange and the bottom purple. If you click on it you’ll see a summary box that says “5-class PuOr”. See the screen shot below for an arrow pointing to the exact location.
![lesson6_screenshot1]()
There’s a lot of other really useful features on the site, including the ability to use only colorblind safe palettes, and you can get HEX and other codes for use in various programs. For now though all we need to remember is the name of our palette, “PuOr”, we can ignore the part that says “5-class”, that’s just letting us know we picked 5 colors from the palette “PuOr”. To read the palette into R for our figures we use the call brewer.pal(). First we specify the number of colors we want, in our case 5, and then the palette, in our case “PuOr”. I’ve saved this to a variable called “cols”. In “cols” I have five different HEX codes for each of my colors. If you look back at the original figure though you’ll see that we actually only need two colors, one for “congruent” data points and one for “incongruent” data points. I originally asked for five colors though because I wanted the darker orange and darker purple from the palette, or the first and fifth colors in the list in “cols”. So now I’m going to save these specific colors to two new variables “col_con” (for congruent trials) and “col_incon” (for incongruent trials). For more details on what the code is doing watch the video. Copy and run the code below putting it above the code for the figure but below the the code for organizing the data.
## SET COLORS FOR FIGURES ####
cols = brewer.pal(5, "PuOr")
col_con = cols[1]
col_incon = cols[5]
Now let’s update our figure to include our new colors. Copy and run the updated code below. Note, in the past when we used scale_fill_manual() our color names were in quotes, here though we don’t use quotes since were calling variables we created earlier.
accuracy.plot = ggplot(data_accuracy_figs, aes(x = half, y = perc_correct,
fill = congruency)) +
geom_boxplot() +
ylim(0, 100) +
geom_hline(yintercept = 50) +
scale_fill_manual(values = c(col_con, col_incon))
pdf("figures/accuracy.pdf")
accuracy.plot
dev.off()
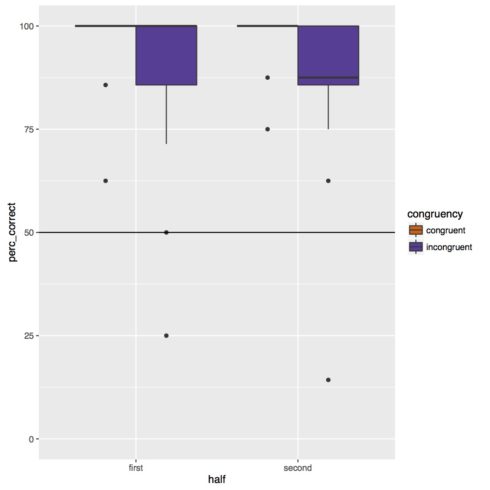
Below is the boxplot. Overall subjects were very accurate on the task, however they did appear to be less accurate on incongruent trials. There does not appear to be any effect of experiment half or any interaction of congruency and experiment half.
![accuracy]()
Now that we have our figure for the accuracy data let’s make our reaction time figure. Go back to the “ORGANIZE DATA” section of the script below the data frame for the accuracy figures. I don’t have to do any summarising since my dependent variable is already continuous (reaction times in milliseconds) but I will update the level labels for “congruency” as I did for the accuracy figure. Copy and run the code below.
# RT data
data_rt_figs = data_rt_clean %>%
mutate(congruency = factor(congruency, levels = c("con", "incon"),
labels = c("congruent", "incongruent")))
Now eventually we’ll make a boxplot, but first we need to make a histogram to be sure the data is normally distributed. Below is the code for the histogram. All of it should be familiar to you at this point. I’ve also included our new colors as fills for the histograms. Copy and run the code below to save the figure to a PDF.
# RT histogram
rt_histogram.plot = ggplot(data_rt_figs, aes(x = rt, fill = congruency)) +
geom_histogram(bins = 30) +
facet_grid(half ~ congruency) +
scale_fill_manual(values = c(col_con, col_incon))
pdf("figures/rt_histogram.pdf")
rt_histogram.plot
dev.off()
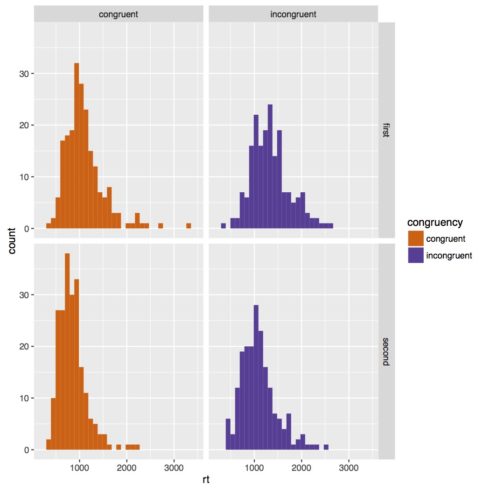
Below is our histogram. Our data looks pretty clearly skewed, so it would probably be good to transform our data to make more normal. It’s actually quite common in reaction time studies to do a log transform of the data, so we’re going to do that.
![rt_histogram]()
To do our transform go back to the cleaning script. We’re going to update the data frame “data_rt_clean” to add a column that does a log 10 transform of our reaction times. Remember, the call log() in R uses log based e, but we’re going to use the call log10() to use log based 10. Copy and run the updated code below.
data_rt_clean = data_accuracy_clean %>%
filter(accuracy == "1") %>%
mutate(rt_log10 = log10(rt))
Now go back to the figures script and rerun the call to make “data_rt_figs”, you don’t need to change anything just rerun it since now “data_rt_clean” has been changed. Okay, now let’s make a new histogram using our log 10 transformed reaction times. Copy and run the code below. It’s the same as our first histogram, just changing the variable for x.
# RT log 10 histogram
rt_log10_histogram.plot = ggplot(data_rt_figs, aes(x = rt_log10, fill = congruency)) +
geom_histogram(bins = 30) +
facet_grid(half ~ congruency) +
scale_fill_manual(values = c(col_con, col_incon))
pdf("figures/rt_log10_histogram.pdf")
rt_log10_histogram.plot
dev.off()
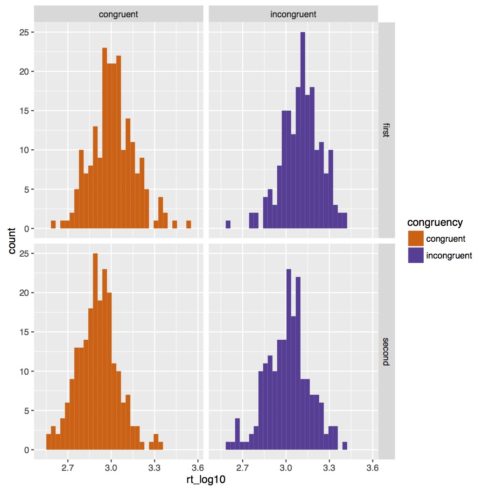
Below is the new histogram. Now our data looks much more normal.
![rt_log10_histogram]()
Now that we have more normal data let’s go ahead and make a boxplot with our log 10 transformed reaction times. All of the code should be familiar to you at this point. Copy and run the code the code below to save the boxplot to a PDF.
# RT log 10 boxplot
rt_log10_boxplot.plot = ggplot(data_rt_figs, aes(x = half, y = rt_log10, fill = congruency)) +
geom_boxplot() +
scale_fill_manual(values = c(col_con, col_incon))
pdf("figures/rt_log10.pdf")
rt_log10_boxplot.plot
dev.off()
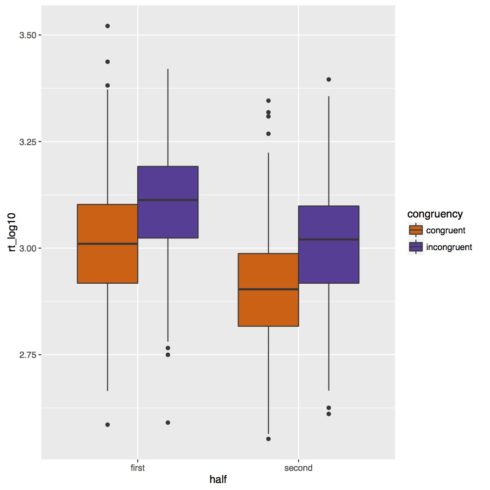
Below is our boxplot of the reaction time data. It looks like we do get a congruency effect such that subjects are slower in incongruent trials than congruent trials. There also appears to be an experiment half effect such that subjects are faster in the second half of the experiment. Based on this figure though it does not look like there is an interaction of congruency and experiment half.
![rt_log10]()
In the script on GitHub you’ll see I’ve added several other parameters to my figures, such as adding a title, customizing how my axes are labeled, and changing where the legend is placed. Play around with those to get a better idea of how to use them in your own figures.
Save your script in the “scripts” folder and use a name ending in “_figures”, for example mine is called “rcourse_lesson6_figures”. Once the file is saved commit the change to Git. My commit message will be “Made figures script.”. Finally, push the commit up to Bitbucket.
Statistics Script (Part 1)
Open a new script. We’ll be using a new package, lme4. Note, the results of the models reported in this lesson are based on version 1.1.12. If you haven’t used this packages before be sure to install it first using the code below. Note, this is a one time call, so you can type the code directly into the console instead of saving it in the script.
install.packages("lme4")
Once you have the package installed, copy the code below to your script and and run it.
## READ IN DATA ####
source("scripts/rcourse_lesson6_cleaning.R")
## LOAD PACKAGES ####
library(lme4)
We’ll also make a header for organizing our data. As for the figures, let’s start with the accuracy data. We aren’t actually going to change anything since all of our variables are already in the right order (“congruent” before “incongruent”, “first” before “second”), so we’ll just set our statistics data frame to our cleaned data frame. Copy and run the code below.
## ORGANIZE DATA ####
# Accuracy data
data_accuracy_stats = data_accuracy_clean
Before we build our model though we need to figure out how to structure our random effects. We decided earlier that subject and item would both be random effects, but just random intercepts? Can we include random slopes? There are different opinions on how you should structure your random effects relative to the amount and type of data you have. I was raised, so to speak, to always start with the maximal random effects structure and then reduce as necessary. (For more information on this method and the reasons for it see this paper by Barr, Levy, Scheepers, & Tily (2013).) Again, other methods are also out there, but this is the one we’ll be using today. You should already know the maximal random effects structure based on your experimental design, but to double check we’ll use xtabs() with “subject_id” and then each of our independent variables. Copy and run the code below.
# Check within or between variables
xtabs(~subject_id+congruency+half, data_accuracy_stats)
If you look at the result for the above call you should see that there are no “0”s in any of the cells. This makes sense, since based on the design of the experiment, every subject saw both congruent and incongruent items, and furthermore they saw each type in each half of the experiment. So, based on this we can include subject as a random slope by the interaction of congruency and experiment half. What about item though? Copy and run the code below.
xtabs(~item+congruency+half, data_accuracy_stats)
Now you should get several cells with “0”s. This also makes sense, as an item is either congruent or incongruent, it can’t be both. However, it’s still possible that an item could appear in each half of the experiment. To check copy and run the code below.
xtabs(~item+half, data_accuracy_stats)
Indeed there are no “0”s, since the design of the experiment was that each item would be repeated once per each block, and since there were four blocks, twice per experiment half per subject. So we can say that we can include item as a random slope by experiment half. Now we can build the actual model. In the code below is our accuracy model. Note, we use glmer() since our dependent variable is accuracy with “0”s and “1”s, so we want a logistic regression. This is also why we have family = "binomial" at the end of the call, same as with glm(). We have our two fixed effects included as main effects and an interaction. We also have random effects for both subject and item, including random slopes in addition to random intercepts as appropriate. Copy and run the code below.
## BUILD MODEL FOR ACCURACY ANALYSIS ####
accuracy.glmer = glmer(accuracy ~ congruency * half +
(1+congruency*half|subject_id) +
(1+half|item), family = "binomial",
data = data_accuracy_stats)
You should have gotten an error message when you ran the model like the one above. This happens when the model doesn’t converge, which means the model gave up before being able to find the best coefficients to fit the data. As a result the coefficients the model gives can’t be trusted, since they are not the best ones, just the ones the model happened to have when it gave up. The main thing to know is do NOT TRUST MODELS THAT DO NOT CONVERGE!!
![lesson6_screenshot2]()
There are a couple different things people will do to try and get a model to converge. If you do a search on the internet you will find various types of advice including increasing the number of iterations or changing the optimizer. What’s most common though is to reduce your random effects structure. Most likely your model is not converging because your model is too complex for the number of data points you have. A good rule of thumb is the more complex the model (interactions in fixed effects, random slopes) the more data points are needed for the model to converge. So, what’s the best way to go about reducing the random effects structure?
Below is a table starting with the maximal random effects structure for something like subject in our model (random intercept and random slope by interaction of two variables) down to the simplest model (random intercept only). This is by no means a gold standard or a field established method, this simply represents what I’ve done in the past when reducing my models. Feel free to use it, but also know that other researchers may have other preferences and methods. As you can see the first reduction is “uncorrelated intercept and slope”. The syntax for adding random slopes we’ve used thus far has been (1+x1|s) where “x1” is one of our fixed effects and “s” is one of our random effects. In this syntax we include both a random intercept and a random slope, and furthermore they are correlated. However, one way to simplify the model is to separate the random intercept and the random slope and uncorrelated them. To do that we first add the random intercept (1|s) and then the random slope (0+x1|s), the use of the “0” makes it uncorrelated with the random slope. The rest of the table goes through further ways to reduce the random effect structure such as by dropping an interaction, or one variable entirely. Note, in the table here I first drop x2 to preserve the slope with x1, and then if it still doesn’t converge try dropping x1 in favor of x2. Which variable you drop first will depend on your data. In general I try to drop the variable that I think matters less or accounts for less of the variance, so as to preserve the variable that I expect to have more variance.
Table: R Code for the Random Effects Structure
| Random Effects Structure |
R Code |
| maximal |
(1+x1*x2|s) |
| uncorrelated intercept and slope |
(1|s) + (0+x1*x2|s) |
| no interaction |
(1+x1+x2|s) |
| uncorrelated intercept and slope, no interaction |
(1|s) + (0+x1+x2|s) |
| no x2 |
(1+x1|s) |
| uncorrelated intercept and slope, no x2 |
(1|s) + (0+x1|s) |
| no x1 |
(1+x2|s) |
| uncorrelated intercept and slope, no x1 |
(1|s) + (0+x2|s) |
| intercept only |
(1|s) |
One final thing this table doesn’t take into account is how to reduce the structure when you have two random effects, such as in our case where we have both subjects and items. Again, I don’t know of any hard and fast rule for how to do this, it seems more a matter of preference. I tend to reduce them in parallel, so for example I’d have (1+x1|s1) + (1+x1|s2) before (1+x1*x2|s1) + (1|s2). However, I still need to pick one to reduce first. I generally go for whichever one I think will have the smaller amount of variance across different iterations, so for example I think that subjects will have more variance than items, so I reduce items first to maintain the more complex structure for subject.
Given this I tried various models until I found one that would converge. I won’t go through each model but simply give the the final model I used that did converge. The model is below. Copy and run the code.
accuracy.glmer = glmer(accuracy ~ congruency * half +
(1|subject_id) +
(0+half|subject_id) +
(1|item), family = "binomial",
data = data_accuracy_stats)
Just as for a simple linear or logistic regression I can use the summary() call to get more information about the model. Copy and run the code below.
# Summarise model and save
accuracy.glmer_sum = summary(accuracy.glmer)
accuracy.glmer_sum
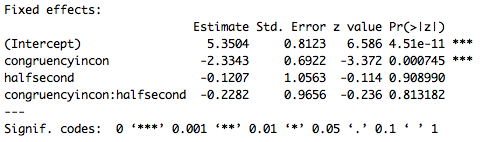
Part of the output of the summary call is below. Remember, this is not an ANOVA, it is a logistic regression, and since there is an interaction in the model we need to interpret the intercept and main effects given the baseline level of each variable. So the intercept is the mean (in logit space) accuracy for congruent trials (the baseline for congruency) in the first half of the experiment (the baseline for experiment half). Furthermore the effect of congruency is specific to the first half of the experiment, and the effect of experiment half is specific to congruent trials. Based on these results it appears that there is an effect of congruency where subjects are less accurate on incongruent trials (specifically in the first half of the experiment). There does not appear to be an effect of experiment half or an interaction. However, these p-values are based on the Wald z statistic which can be biased for small sample sizes, and give overly, generously low p-values. As a result in the next lesson we’ll go over a different way to compute p-values for LMEMs.
![lesson6_screenshot3]()
In addition to getting the summary() the model we can also look at the coefficients of the random effects with the coef() call. Copy and run the code below.
# Get coefficients and save
accuracy.glmer_coef = coef(accuracy.glmer)
accuracy.glmer_coef
Below are the coefficients for item. Note, the coefficients for subjects are also available in the full call. Interpreting these can be complicated (particularly because of the structure for the random effect of subject), but one thing I want to draw your attention to is that for the intercept each item has a different value, this is because we added a random intercept of item, while there is no difference for the variables or the interactions of the variables, since no random slopes were added.
![lesson6_screenshot4]()
Now let’s do the same thing for our analysis of reaction times. Start by going back to the top of the script at the bottom of the “ORGANIZE DATA” section and copy and run the following to make our data for the reaction time analysis.
# RT data
data_rt_stats = data_rt_clean
Again, I’ll start with the model with maximal random effects structure. The maximal random effects structure is the same as for the accuracy model. If you would like to double check feel free to rerun the xtabs() calls using “data_rt_stats” instead of “data_accuracy_stats”. For this model we use lmer() since our dependent variable is continuous so a linear model is appropriate; as a result we also don’t need the family = "binomial" call at the end. Also, remember we’re using log 10 reaction times as our dependent variable. Copy and run the code below when you’re ready.
## BUILD MODEL FOR REACTION TIME ANALYSIS ####
rt_log10.lmer = lmer(rt_log10 ~ congruency * half +
(1+congruency*half|subject_id) +
(1+half|item),
data = data_rt_stats)
Unlike for the accuracy analysis the maximal model should have converged for you, so there’s no need to reduce the random effects structure. Copy and run the code below to get the summary of the model.
# Summarise model and save
rt_log10.lmer_sum = summary(rt_log10.lmer)
rt_log10.lmer_sum
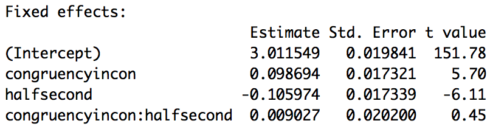
Part of the output of the summary call is below. Unlike for the logistic regression we don’t get any p-values, all the more reason to use the method to be discussed in Part 2 of this lesson. However, a general rule of thumb is if the t-value has an absolute value of 2 or greater it will be significant at p < 0.05. Based on these results then subjects are slower on incongruent trials in the first half of the experiment (don’t forget baselines!) and faster in the second half of the experiment for congruent items. There is no interaction of congruency and half. So, while for the accuracy analysis we got an effect of congruency but no effect of experiment half, for the reaction time analysis we get an effect of both congruency and experiment half.
![lesson6_screenshot5]()
Finally, we can also look at the coefficients of the random effects. Copy and run the code below.
# Get coefficients and save
rt_log10.lmer_coef = coef(rt_log10.lmer)
rt_log10.lmer_coef
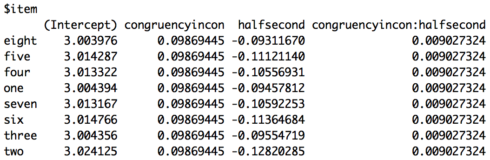
The coefficients for item are below. As you can see they look a little different from the other model. One reason is because we now have our random intercept and our random slope. However, it’s still the case that each item has the same values for congruency, but now also different values for experiment half.
![lesson6_screenshot6]()
You’ve now run two LMEMs, one with logistic regression and one with linear regression, in R! Save your script in the “scripts” folder and use a name ending in “_pt1_statistics”, for example mine is called “rcourse_lesson6_pt1_statistics”. Once the file is saved commit the change to Git. My commit message will be “Made part 1 statistics script.”. Finally, push the commit up to Bitbucket. If you are done with the lesson you can go to your Projects menu and click “Close Project”.
Conclusion and Next Steps
Today you learned how to run an LMEM, giving us the power to look at multiple types of effects within a single model and not have to worry about unbalanced data. You were also introduced to the packages RColorBrewer and lme4, and as always expanded your knowledge of dplyr and ggplot2 calls. However, there are a couple concerns you may have that still makes an ANOVA more attractive. For example, our baseline issue is back, and we don’t seem to have any straightforward p-values, which, while shouldn’t be that important, would be nice to have. In the second half of this lesson both of these issues will be addressed.
* From here on out I’ll be using the word “subject” instead of “participant”. While “participant” is the more appropriate word to use in any kind of write-up, I’ve found people are more used to “subject” when discussing data and analyses.
Related Post
- R for Publication by Page Piccinini: Lesson 6, Part 2 – Linear Mixed Effects Models
- Cross-Validation: Estimating Prediction Error
- Interactive Performance Evaluation of Binary Classifiers
- Predicting wine quality using Random Forests
- Bayesian regression with STAN Part 2: Beyond normality